We are focused on using high-resolution NMR spectroscopy as a tool to better comprehend the mechanisms of signal transmission in the brain and disease transmission through the stomach. In particular, we are interested in understanding how protein structure, protein motions and their biophysical properties impact and influence the biological function of these proteins.
Our lab is currently investigating two chaperone proteins and two families of signaling proteins:
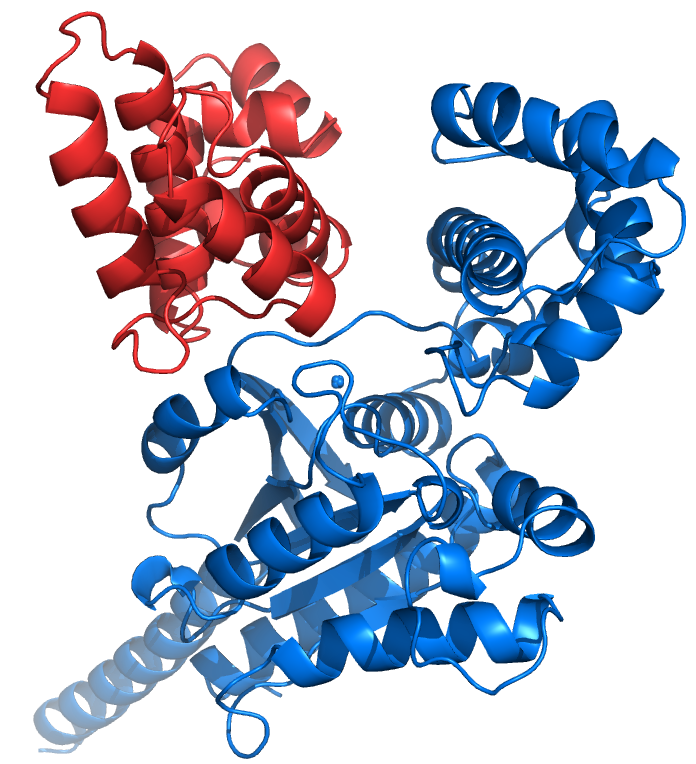
HdeA and HdeB
The stomach is an important barricade that helps to kill many bacteria before they can cause illness, in part by using its acidity to inactivate bacterial proteins. Some bacteria contain small chaperone proteins called HdeA and HdeB that help protect other proteins from becoming permanently inactivated and therefore help bacteria survive and ultimately cause infection. We have done studies showing that HdeA unfolds below pH 2.6 and have mapped the structural details of its unfolding and activation. However, there has been a lack of data that monitors, in detail, the chaperoning mechanism of HdeA and HdeB.
We are using NMR spectroscopy as our primary tool to investigate these mechanisms at atomic resolution, allowing us to get a comprehensive picture of the interactions between HdeA, HdeB and their client proteins. This work is moving us closer to the detailed understanding required for drug designers to target HdeA or HdeB and combat dysentery, a disease that infects millions of people worldwide and is often fatal.
RGS proteins and their Galpha targets
RGS proteins are responsible for turning off a common type of signalling cascade in the brain. Ultimately, RGS proteins control the relay of signals that are triggered by light, smell, hormones or neurotransmitters. Their primary binding targets are the alpha subunits of G proteins.
We would like to understand how proteins in both of these families bind to their specific targets (and avoid other targets that are only slightly different) and how changes in protein structure help in transmitting signals. Since both protein families are critical for the initiation or regulation of signalling cascades, our research may lead to increased understanding of the causes of conditions such as cerebral stroke, chronic pain, cancer, epilepsy, depression, schizophrenia and neurodegenerative diseases (including Alzheimer’s and Parkinson’s diseases) and can aid in the design of drugs to treat these conditions.
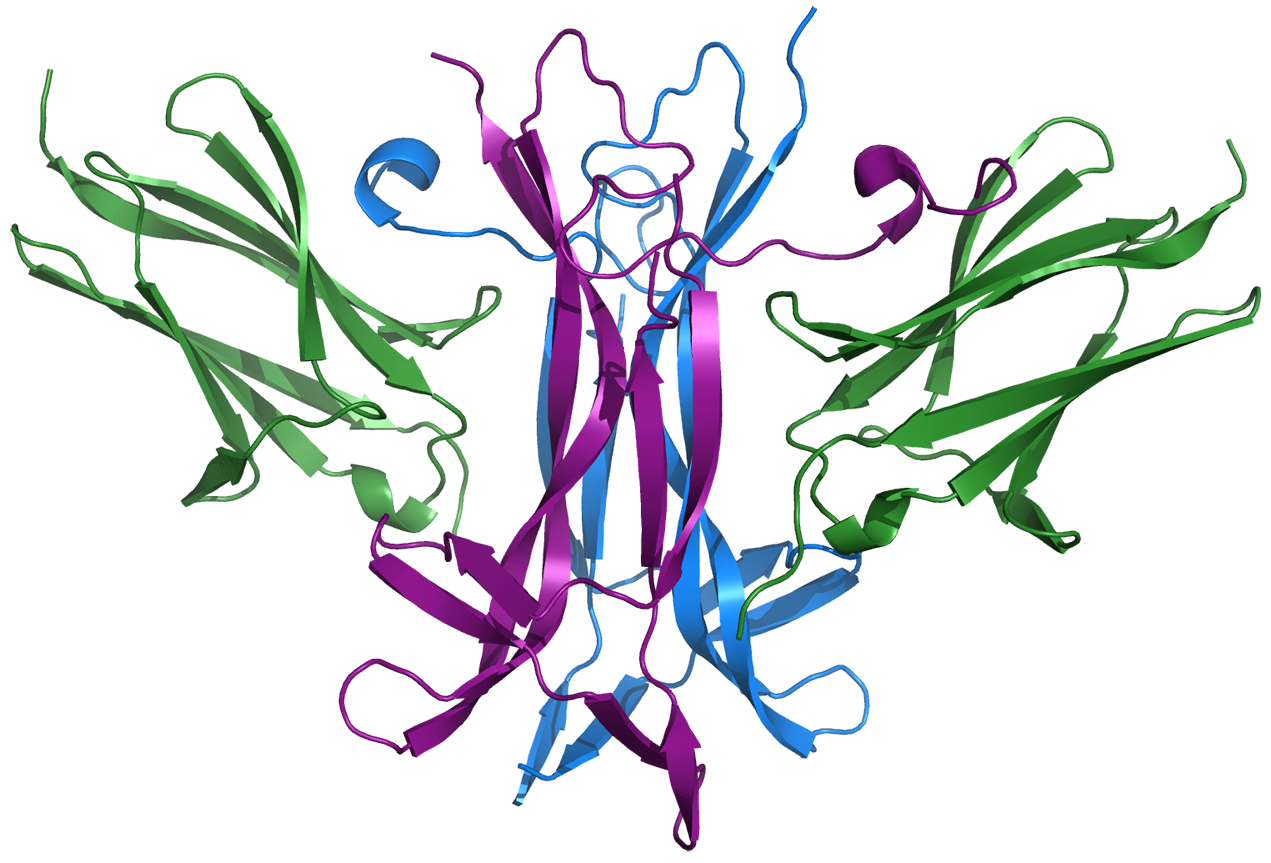
Neurotrophins and their Trk receptors
Neurotrophins are responsible for providing the signal for the growth and differentiation of neurons in the brain. These proteins also have an important influence on synaptic function, learning and memory. They have two receptors: p75 (which binds all neurotrophins with equal affinity) and Trk receptors (each of which is selective for one or two neurotrophins). We are most interested in studying neurotrophin-Trk interactions.
Lab and investigative methods
In the lab we work to express our proteins of interest using recombinant techniques and purify the isotopically labeled samples. We then take our samples upstairs to run our NMR experiments on our high-resolution 600 MHz NMR spectrometer (“Agnes”), which was installed in September 2011. Occasionally we perform some of our experiments off-campus when we need a higher-field spectrometer. The protein NMR spectroscopic techniques used to address these biological questions are, by necessity, more sophisticated and provide significantly more information than can be obtained by recording the standard one-dimensional 1H or 13C spectra. Our experiments require the recording of 2D and 3D spectra, in which the signals of multiple nuclei are linked to each other to provide detailed information about structural and dynamic properties of proteins.
Anyone interested in working in the lab will therefore learn a wide variety of wetlab techniques and will learn how to analyze 2D and 3D multinuclear NMR spectra to obtain information about the structure, motions and interactions of our proteins.